gene view
Abstract
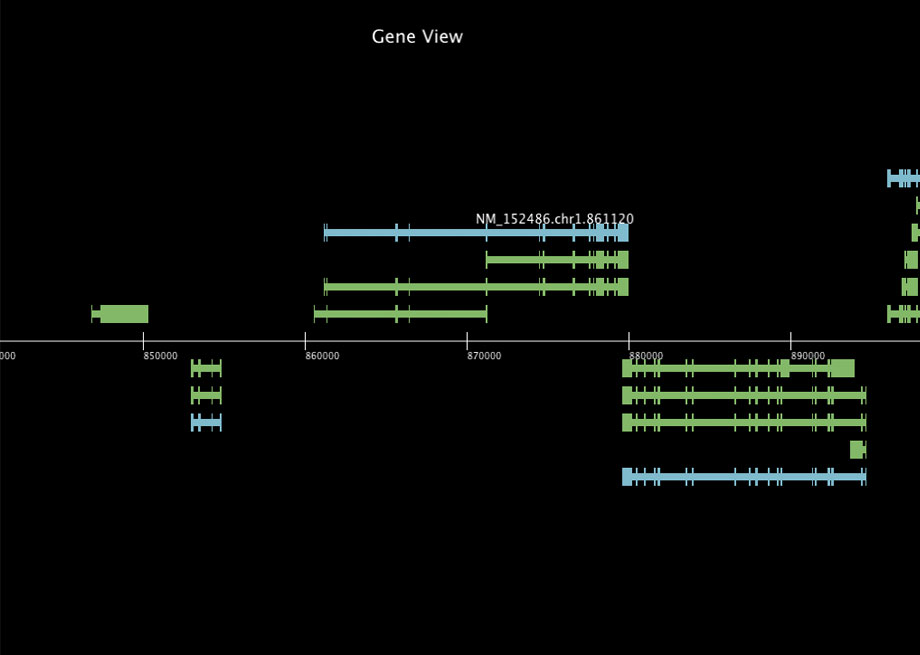
"gene view" is a viewer for the Human genome data (Human Genome, UCSC hg.19) with accessing to the DAS (The Distributed Annotation System) server in UCSC. This gallery shows visualization for data of refGene and knowGene that can get with "type" command of the DAS protocol.
This viewer shows the exon-intron structures and you can browse data with your mouse drag. When you drag your mouse horizontally, the shown area of genome will be moved. When you drag your mouse vertically, the zoom level of genome will be changed. When you mouse-over an object, you can see the annotation (name) of the object.
https://github.com/casmi/gene-view
Git Repository
https://github.com/casmi/gene-view.git
Inside of Program
Code
- It automatically get the data from the UCSC Human annotation DAS server and stores to a local machine cache (GeneView.java, GeneLoader.java).
- The data which is XML format is read and parsed, and is stored to objects (GeneXMLParser.java, Exon.java, Gene.java).
- After that, the data is set to the appropriate position according to the base pairs (GeneView.java).
- To draw the data, it uses the Rect class for each element (GeneElement.java).
- It also uses MouseOverCallback to objects (GeneElement.java). When the mouse is on the objects, it shows the annotation name: refGene name or knownGene name (GeneView.java).
- It also display the scale (basepairs) to show which area is shown.

